Classes | |
| class | OBCisTransStereo |
| Class for handling and storing cis/trans stereochemistry. More... | |
| class | OBSquarePlanarStereo |
| Class for handling and storing square planar stereochemistry. More... | |
| class | OBStereo |
| Placeholder for enums & Ref/Refs related functions. More... | |
| struct | OBStereoUnit |
| Struct representing a single stereogenic unit. More... | |
| class | OBStereoBase |
| Base class for all stereochemistry classes. More... | |
| class | OBStereoFacade |
| Facade to simplify retrieval of OBStereoBase derived objects. More... | |
| class | OBTetrahedralStereo |
| Class for handling and storing tetrahedral atom stereochemistry. More... | |
| class | OBTetraNonPlanarStereo |
| Base class for handling and storing non-planar stereochemistry with 4 reference atom ids. More... | |
Typedefs | |
| typedef std::vector< OBStereoUnit > | OBStereoUnitSet |
| typedef std::vector < OBStereoUnitSet > | OBStereoUnitSetOfSets |
Functions | |
| ostream & | operator<< (ostream &out, const OpenBabel::OBCisTransStereo &ct) |
| ostream & | operator<< (ostream &out, const OpenBabel::OBCisTransStereo::Config &cfg) |
| ostream & | operator<< (ostream &out, const OpenBabel::OBSquarePlanarStereo &ct) |
| ostream & | operator<< (ostream &out, const OpenBabel::OBSquarePlanarStereo::Config &cfg) |
| ostream & | operator<< (ostream &out, const OpenBabel::OBTetrahedralStereo &ts) |
| ostream & | operator<< (ostream &out, const OpenBabel::OBTetrahedralStereo::Config &cfg) |
High level functions | |
| void | PerceiveStereo (OBMol *mol, bool force=false) |
| void | StereoFrom2D (OBMol *mol, std::map< OBBond *, enum OBStereo::BondDirection > *updown=NULL, bool force=false) |
| void | StereoFrom3D (OBMol *mol, bool force=false) |
| void | StereoFrom0D (OBMol *mol) |
Low level functions | |
| std::vector < OBTetrahedralStereo * > | TetrahedralFrom3D (OBMol *mol, const OBStereoUnitSet &stereoUnits, bool addToMol=true) |
| std::vector < OBTetrahedralStereo * > | TetrahedralFrom2D (OBMol *mol, const OBStereoUnitSet &stereoUnits, bool addToMol=true) |
| std::vector < OBTetrahedralStereo * > | TetrahedralFrom0D (OBMol *mol, const OBStereoUnitSet &stereoUnits, bool addToMol=true) |
| std::vector< OBCisTransStereo * > | CisTransFrom3D (OBMol *mol, const OBStereoUnitSet &stereoUnits, bool addToMol=true) |
| std::vector< OBCisTransStereo * > | CisTransFrom2D (OBMol *mol, const OBStereoUnitSet &stereoUnits, const std::map< OBBond *, enum OBStereo::BondDirection > *updown=NULL, bool addToMol=true) |
| bool | TetStereoToWedgeHash (OBMol &mol, std::map< OBBond *, enum OBStereo::BondDirection > &updown, std::map< OBBond *, OBStereo::Ref > &from) |
| std::set< OBBond * > | GetUnspecifiedCisTrans (OBMol &mol) |
| void | StereoRefToImplicit (OBMol &mol, OBStereo::Ref atomId) |
| std::vector< OBCisTransStereo * > | CisTransFrom0D (OBMol *mol, const OBStereoUnitSet &stereoUnits, bool addToMol=true) |
Stereogenic unit identification | |
| OBStereoUnitSet | FindStereogenicUnits (OBMol *mol, const std::vector< unsigned int > &symClasses) |
| OBStereoUnitSet | FindStereogenicUnits (OBMol *mol, const std::vector< unsigned int > &symClasses, const Automorphisms &automorphisms) |
There are many molecules which contain stereogenic elements. However, certain cases (i.e. tetrahedral, cis/trans) are more common than others (i.e. allene, biphenyl, octrahedral, ...). For the common stereogenic units, classes are provided. The inheritance of these classes resembles the way they are split into groups.
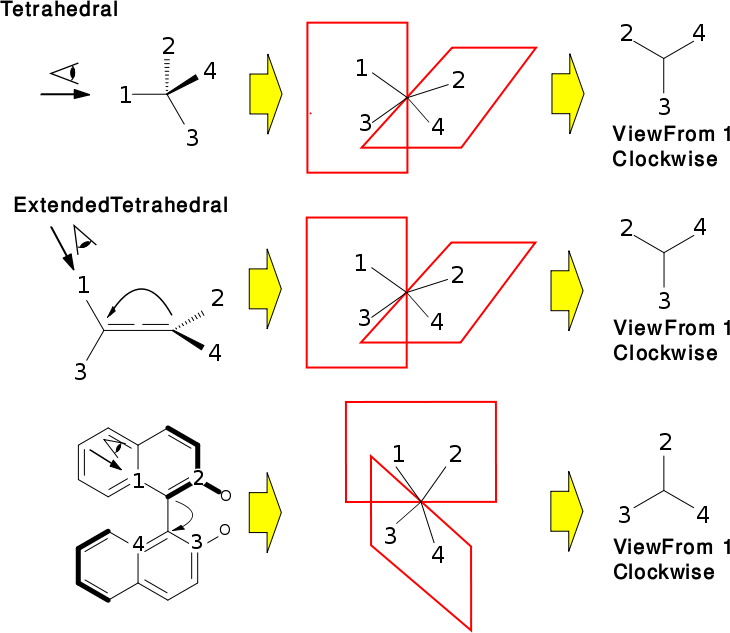
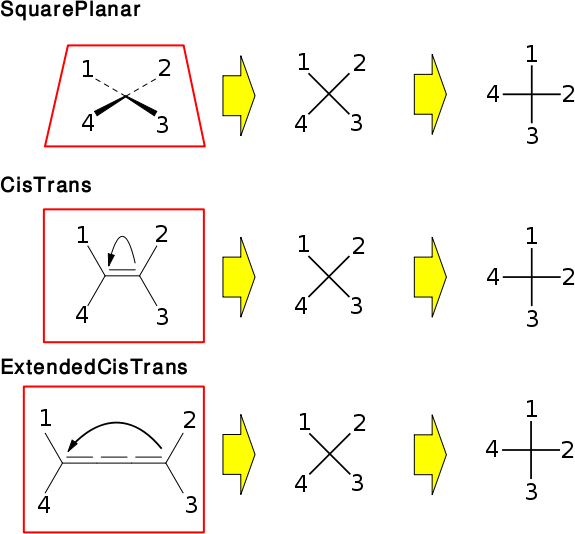
All specific classes (i.e. OBTetrahedralStereo, ...) have embedded Config structs which define the actual stereochemistry. All these Config structs use OBStereo::Ref values to reference or uniquely identify atoms. Make sure to read about OBStereo::Ref and the related functions (in OBStereo). OBStereo is also a placeholder for various enums with predefined values for parameters etc. These enums are used throughout the different stereo classes but having these enums in a single location makes it easier to remember. When working with stereo classes, you normally don't need to use any of the parent classes directly. Only OBStereo and the specific class are needed.
The OBStereoFacade hides the complexity of working with stereochemistry. When using openbabel as a library, this is by far the easiest way to access stereochemistry information. The header for the specific OBStereo::Type type is all you need to include. These are:
All these headers also include openbabel/stereo/stereo.h providing declarations for OBStereo & OBStereoFacade.
#include <iostream> #include <openbabel/mol.h> #include <openbabel/obconversion.h> #include <openbabel/stereo/tetrahedral.h> using namespace OpenBabel; int main() { OBMol mol; OBConversion conv; conv.SetInFormat("smi"); conv.ReadString(&mol, "C[C@H](Cl)Br"); OBStereoFacade facade(&mol); FOR_ATOMS_OF_MOL(atom, mol) { if (facade.HasTetrahedralStereo(atom->GetId())) std::cout << facade.GetTetrahedralStereo(atom->GetId()) << std::endl; } }
All specific stereo classes and their embedded Config struct have an operator<< function which allows them to be used with std::ostream objects (e.g. std::cout, std::err, ...). These functions are often useful when debugging code.
The detection of stereogenic units start with symmetry analysis. However, a complete symmetry analysis also needs to take stereochemistry into account. In practice, this means stereochemistry will be found iteratively. At each iteration, the current atom symmetry classes are used to identify stereogenic units. The details about how the symmetry classes are used depends on the type (OBStereo::Type) of stereogenic unit. For tetrahedral centers, having 3 heavy atom neighbors with different symmetry classes or 4 neighbors with different symmetry classes means the atom is chiral. See FindStereogenicUnits() for details.
After identifying the stereogenic units, Config structs with all the information on the spacial arrangement of the groups still have to be created. This involves interpreting various ways to represent stereochemisrty:
Both StereoFrom3D() and StereoFrom2D() delete all existing stereochemistry objects before adding new ones. For molecules with 3D coordinates, it is evident that all information is specified by the coordinates itself. However, if a file format uses stereo parity flags, Config structs must be constructed using lower level functions and StereoFrom3D() should not be called. In these cases information could be lost by calling StereoFrom3D() after reading the file (the stereo flag might have indicated the stereochemistry was unspecified or the flag might not match the coordinates). In the case of 2D molecules, the coordinates together with bond properties (OBBond::Hash, OBBond::Wedge, OBBond::WedgeOrHash and OBBond::CisOrTrans) define the stereochemistry. Again, lower level functions can be used when stereo flags need to be used.
StereoFrom0D() works slightly different than 3D/2D. Here, deleting the stereochemistry would always result in lost information. Instead StereoFrom0D() only adds new objects for stereogenic units which were previously not found. For example, a smiles is read which has two tetrahedral centers. Only one has stereochemistry specified using a '@' character. StereoFrom0D() will detect the second tetrahedral atom and add an OBTetrahedralStereo object to the molecule. The Config::specified flag for the newly added structs is always set to false.
Assuming the format code has correctly set the molecule dimensions (OBMol::GetDimesions), PerceiveStereo() will automatically select the correct function to call. When StereoFrom3D(), StereoFrom2D() or StereoFrom0D() are not used, make sure to always set OBMol::HasChiralityPerceived() before returning from the format's ReadMolecule().
For many file formats no additional code is needed. For example, if a 3D format doesn't require stereo parity flags, writing the coordinates is enough. For 2D file formats it will often suffice to write the coordinates and bond properties. If parity flags are needed, the OBStereoFacade class can be used to retreive the objects for all types of stereochemistry supported by the file format.
| typedef std::vector<OBStereoUnit> OBStereoUnitSet |
A single set of OBStereoUnit objects.
This type can be used to represent all stereogenic units in a molecule and is used as return type of FinStereogenicUnits(). This set is also the input for many functions requiring this information (e.g. StereoFrom2D, ...).
| typedef std::vector<OBStereoUnitSet> OBStereoUnitSetOfSets |
A set of sets of OBStereoUnit objects.
This type is used for cases where there is some relationship between individual OBStereoUnit objects.
| ostream& std::operator<< | ( | ostream & | out, |
| const OpenBabel::OBCisTransStereo & | ct | ||
| ) |
OBCisTransStereo::Config cfg; cfg.begin = 0; cfg.end = 1; cfg.refs = OBStereo::MakeRefs(2, 3, 4, 5); cfg.shape = OBStereo::ShapeU; OBCisTransStereo ct(mol); ct.SetConfig(cfg) cout << "ct = " << ct << endl; // output OBCisTransStereo(begin = 0, end = 1, refs = 2 3 4 5, shape = U)
| ostream& std::operator<< | ( | ostream & | out, |
| const OpenBabel::OBCisTransStereo::Config & | cfg | ||
| ) |
OBCisTransStereo::Config cfg; cfg.begin = 0; cfg.end = 1; cfg.refs = OBStereo::MakeRefs(2, 3, 4, 5); cfg.shape = OBStereo::ShapeU; cout << "cfg = " << cfg << endl; // output OBCisTransStereo::Config(begin = 0, end = 1, refs = 2 3 4 5, shape = U)
| ostream& std::operator<< | ( | ostream & | out, |
| const OpenBabel::OBSquarePlanarStereo & | ct | ||
| ) |
OBSquarePlanarStereo::Config cfg; cfg.center = 0; cfg.refs = OBStereo::MakeRefs(1, 2, 3, 4); cfg.shape = OBStereo::ShapeU; OBSquarePlanarStereo ct(mol); ct.SetConfig(cfg) cout << "ct = " << ct << endl; // output OBSquarePlanarStereo(center = 0, refs = 1 2 3 4, shape = U)
| ostream& std::operator<< | ( | ostream & | out, |
| const OpenBabel::OBSquarePlanarStereo::Config & | cfg | ||
| ) |
OBSquarePlanarStereo::Config cfg; cfg.center = 0; cfg.refs = OBStereo::MakeRefs(1, 2, 3, 4); cfg.shape = OBStereo::ShapeU; cout << "cfg = " << cfg << endl; // output OBSquarePlanarStereo::Config(center = 0, refs = 1 2 3 4, shape = U)
| void OpenBabel::PerceiveStereo | ( | OBMol * | mol, |
| bool | force = false |
||
| ) |
Convert 0D/2D/3D coordinates to OBStereo objects. The right function will be selected based on the molecule's dimensionality (i.e. OBMol::GetDimension()).
| void OpenBabel::StereoFrom2D | ( | OBMol * | mol, |
| std::map< OBBond *, enum OBStereo::BondDirection > * | updown = NULL, |
||
| bool | force = false |
||
| ) |
Convert the 2D depiction of molecule mol to OBStereo objects. This function makes use of the lower level functions TetrahedralFrom2D(), CisTransFrom2D(), SquarePlanarFrom2D(), ...
First, symmetry analysis taking stereochemistry into account is performed iteratively (see OBGraphSym). Next the 2D coordinates, OBBond::Wedge, OBBond::Hash, OBBond::WedgeOrHash and OBBond::CisOrTrans are used to construct OBStereoBase derived objects to store the stereochemistry. These objects will be added to mol.
Unless perception is forced, this function does nothing if stereochemistry has already been perceived (i.e. OBMol::HasChiralityPerceived()). Before doing the actual perception, any data of the OBGenericDataType::StereoData type will be deleted.
Reference:
[1] T. Cieplak, J.L. Wisniewski, A New Effective Algorithm for the
Unambiguous Identification of the Stereochemical Characteristics of
Compounds During Their Registration in Databases. Molecules 2000, 6,
915-926, http://www.mdpi.org/molecules/papers/61100915/61100915.htm
| mol | The molecule containing 2D coordinates. |
| updown | A map of OBStereo::BondDirection for cis/trans bonds |
| force | Force to run the perception even if the results are cached. |
| void OpenBabel::StereoFrom3D | ( | OBMol * | mol, |
| bool | force = false |
||
| ) |
Convert the 3D coordinates of molecule mol to OBStereo objects. This function makes use of the lower level functions TetrahedralFrom3D(), CisTransFrom3D(), SquarePlanarFrom3D(), ...
Unless perception is forced, this function does nothing if stereochemistry has already been perceived (i.e. OBMol::HasChiralityPerceived()). Before doing the actual perception, any data of the OBGenericDataType::StereoData type will be deleted.
| mol | The molecule containing 3D coordinates. |
| force | Force to run the perception even if the results are cached. |
| void OpenBabel::StereoFrom0D | ( | OBMol * | mol ) |
Add missing OBStereo objects. Unlike StereoFrom3D() and StereoFrom2D(), this method only adds objects for previously unidentified objects since we don't want to loose any information. The Config::specified flag for the newly added structs is always set to false.
For example, a smiles is read which has two tetrahedral centers. Only one has stereochemisrty specified using a '@' character. StereoFrom0D() will detect the second tetrahedral atom and add an OBTetrahedralStereo object to the molecule.
| mol | The molecule. |
| std::vector<OBTetrahedralStereo*> OpenBabel::TetrahedralFrom3D | ( | OBMol * | mol, |
| const OBStereoUnitSet & | stereoUnits, | ||
| bool | addToMol = true |
||
| ) |
Get a vector with all OBTetrahedralStereo objects for the molecule. This function is used by StereoFrom3D() with the addToMol parameter is set to true.
The algorithm to convert the 3D coordinates to OBTetrahedralStereo object uses the sign of the volume described by the 4 center atom neighbors. Given 4 points 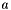 ,
, 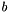 ,
, 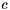 and
and 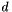 , the signed volume
, the signed volume 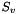 is defined as:
is defined as:
![\[ S_v = \left| \begin{array}{ccc} x_b - x_a & y_b - y_a & z_b - z_a \\ x_c - x_a & y_c - y_a & z_c - z_a \\ x_d - x_a & y_d - y_a & z_d - z_a \end{array} \right| \]](form_5.png)
The sign of 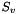 changes when any of the points cross the plane defined by the other 3 points. To make this less abstract one could say that a change of sign is equal to inverting the tetrahedral stereochemistry.
changes when any of the points cross the plane defined by the other 3 points. To make this less abstract one could say that a change of sign is equal to inverting the tetrahedral stereochemistry.
In case there are only 3 neighbor atoms for the tetrahedral center, the center atom itself is used as 4th point. This only changes the magnitude and not the sign of 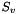 because the center atom is still on the same side of the plane.
because the center atom is still on the same side of the plane.
This function is also used for symmetry analysis to handle cases where there are two atoms in the same symmetry class that don't have the same stereochemistry. In this situation, the addToMol parameter is set to false and the returned objects will need to be deleted explicitly.
| mol | The molecule. |
| stereoUnits | The stereogenic units. |
| addToMol | If true, the OBTetrahedralStereo objects will be added to the molecule using OBBase::SetData(). |
Referenced by OpenBabel::CanonicalLabels(), and OBBuilder::CorrectStereoAtoms().
| std::vector<OBTetrahedralStereo*> OpenBabel::TetrahedralFrom2D | ( | OBMol * | mol, |
| const OBStereoUnitSet & | stereoUnits, | ||
| bool | addToMol = true |
||
| ) |
Get a vector with all OBTetrahedralStereo objects for the molecule. This function is used by StereoFrom2D() with the addToMol parameter is set to true.
The algorithm to convert the 2D coordinates and bond properties (i.e. OBBond::Wedge, OBBond::Hash, OBBond::WedgeOrHash and OBBond::CisOrTrans) uses the sign of a triangle. Given 3 points 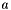 ,
, 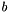 and
and 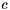 , the sign of the trianle
, the sign of the trianle 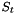 is defined as:
is defined as:
![\[ S_t = (x_a - x_c) (y_b - y_c) - (y_a - y_c) (x_b - x_c) \]](form_7.png)
This is equation 6 from on the referenced web page. The 3 points used to calculate the triangle sign always remain in the same plane (i.e. z = 0). The actual meaning of 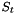 (i.e. assignment of OBStereo::Winding) depends on the 4th atom. When the atom is in front of the plane, the sign should be changed to have the same absolute meaning for an atom behind the plane and the same triangle. It is important to note that none of the z coordinates is ever changed, the molecule always stays 2D (unlike methods which set a pseudo-z coordinate).
(i.e. assignment of OBStereo::Winding) depends on the 4th atom. When the atom is in front of the plane, the sign should be changed to have the same absolute meaning for an atom behind the plane and the same triangle. It is important to note that none of the z coordinates is ever changed, the molecule always stays 2D (unlike methods which set a pseudo-z coordinate).
This function is also used for symmetry analysis to handle cases where there are two atoms in the same symmetry class that don't have the same stereochemistry. In this situation, the addToMol parameter is set to false and the returned objects will need to be deleted explicitly.
Reference:
[1] T. Cieplak, J.L. Wisniewski, A New Effective Algorithm for the
Unambiguous Identification of the Stereochemical Characteristics of
Compounds During Their Registration in Databases. Molecules 2000, 6,
915-926, http://www.mdpi.org/molecules/papers/61100915/61100915.htm
| mol | The molecule. |
| stereoUnits | The stereogenic units. |
| addToMol | If true, the OBTetrahedralStereo objects will be added to the molecule using OBBase::SetData(). |
Referenced by OpenBabel::CanonicalLabels().
| std::vector<OBTetrahedralStereo*> OpenBabel::TetrahedralFrom0D | ( | OBMol * | mol, |
| const OBStereoUnitSet & | stereoUnits, | ||
| bool | addToMol = true |
||
| ) |
Get a vector with all OBTetrahedralStereo objects for the molecule. This function is used by StereoFrom0D() with the addToMol parameter is set to true. There is no algorithm used here, all specified flags will be set to false.
This function is also used for symmetry analysis to handle cases where there are two atoms in the same symmetry class that don't have the same stereochemistry. In this situation, the addToMol parameter is set to false and the returned objects will need to be deleted explicitly.
| mol | The molecule. |
| stereoUnits | The stereogenic units. |
| addToMol | If true, the OBTetrahedralStereo objects will be added to the molecule using OBBase::SetData(). |
Referenced by OpenBabel::CanonicalLabels().
| std::vector<OBCisTransStereo*> OpenBabel::CisTransFrom3D | ( | OBMol * | mol, |
| const OBStereoUnitSet & | stereoUnits, | ||
| bool | addToMol = true |
||
| ) |
Get a vector with all OBCisTransStereo objects for the molecule. This function is used by StereoFrom3D() with the addToMol parameter is set to true.
The algorithm to convert the 3D coordinates to OBCisTransStereo objects considers the signed distance between the attached atoms and the plane through the double bond at right angles to the plane of the attached atoms. Bonds on the same side (cis) will share the same sign for the signed distance.
Missing atom coordinates (OBStereo::ImplicitRef) and their bond vectors will be computed if needed.
0 3 Get signed distance of 0 and 2 to the plane
\ / that goes through the double bond and is at
C==C right angles to the stereo bonds.
/ \
1 2 If the two signed distances have the same sign
then they are cis; if not, then trans.
This function is also used for symmetry analysis to handle cases where there are two atoms in the same symmetry class that don't have the same stereochemistry. In this situation, the addToMol parameter is set to false and the returned objects will need to be deleted explicitly.
| mol | The molecule. |
| stereoUnits | The stereogenic units. |
| addToMol | If true, the OBCisTransStereo objects will be added to the molecule using OBBase::SetData(). |
Referenced by OpenBabel::CanonicalLabels(), and OBBuilder::CorrectStereoBonds().
| std::vector<OBCisTransStereo*> OpenBabel::CisTransFrom2D | ( | OBMol * | mol, |
| const OBStereoUnitSet & | stereoUnits, | ||
| const std::map< OBBond *, enum OBStereo::BondDirection > * | updown = NULL, |
||
| bool | addToMol = true |
||
| ) |
Get a vector with all OBCisTransStereo objects for the molecule. This function is used by StereoFrom2D() with the addToMol parameter is set to true.
This function is also used for symmetry analysis to handle cases where there are two atoms in the same symmetry class that don't have the same stereochemistry. In this situation, the addToMol parameter is set to false and the returned objects will need to be deleted explicitly.
The algorithm for converting the 2D coordinates uses the same triangle sign as TetrahedralFrom2D(). Depending on sign of 2 triangles, the right OBStereo::Shape is selected.
0 3
\ / 2 triangles: 0-1-b & 2-3-a
a==b --> same sign: U
/ \ opposite sign: Z
1 2
| mol | The molecule. |
| stereoUnits | The stereogenic units. |
| updown | A map of OBStereo::BondDirection for cis/trans bonds |
| addToMol | If true, the OBCisTransStereo objects will be added to the molecule using OBBase::SetData(). |
Referenced by OpenBabel::CanonicalLabels().
| bool OpenBabel::TetStereoToWedgeHash | ( | OBMol & | mol, |
| std::map< OBBond *, enum OBStereo::BondDirection > & | updown, | ||
| std::map< OBBond *, OBStereo::Ref > & | from | ||
| ) |
Convert a molecule's OBTetrahedralStereo objects to a series of hash or wedge bonds. Note that the molecule itself is not modified; the result is returned in the maps updown and from, which indicate the origin and direction of each hash or wedge bond.
When converting, the following guidelines are followed when trying to find the best candidate bond to set up/down for each OBTetrahedralStereo object:
If no bond can be found that matches rules 1 and 2 (and in theory this is possible) then an error message is logged and the function returns false. (If you find an example where this occurs, please file a bug.)
| mol | The molecule. |
| updown | A map of OBStereo::BondDirection for each hash/wedge bond |
| from | A map of OBStereo::Ref indicating the origin of each hash/wedge bond |
| std::set<OBBond*> OpenBabel::GetUnspecifiedCisTrans | ( | OBMol & | mol ) |
Return a set of double bonds corresponding to the OBCisTransStereo objects for which the stereochemistry is undefined.
Note that this functions just iterates over the existing OBCisTransStereo objects - it does not try to identify new ones.
| mol | The molecule |
| void OpenBabel::StereoRefToImplicit | ( | OBMol & | mol, |
| OBStereo::Ref | atomId | ||
| ) |
Convert any reference to atomId in a stereo object to an OBStereo::ImplicitRef. This function is called from OBMol::DeleteHydrogens() (via OBMol::DeleteHydrogen()) to remove any explicit references to a hydrogen atom that has been deleted. However, the code is not specific to hydrogen atoms and could be used for other atoms.
| mol | The molecule |
| atomId | The Id of the atom to be converted to an OBStereo::ImplicitRef |
Referenced by OBMol::DeleteHydrogen().
| std::vector<OBCisTransStereo*> OpenBabel::CisTransFrom0D | ( | OBMol * | mol, |
| const OBStereoUnitSet & | stereoUnits, | ||
| bool | addToMol = true |
||
| ) |
Get a vector with all OBCisTransStereo objects for the molecule. This function is used by StereoFrom0D() with the addToMol parameter is set to true. There is no algorithm used here, all specified flags will be set to false.
This function is also used for symmetry analysis to handle cases where there are two atoms in the same symmetry class that don't have the same stereochemistry. In this situation, the addToMol parameter is set to false and the returned objects will need to be deleted explicitly.
| mol | The molecule. |
| stereoUnits | The stereogenic units. |
| addToMol | If true, the OBCisTransStereo objects will be added to the molecule using OBBase::SetData(). |
Referenced by OpenBabel::CanonicalLabels().
| OBStereoUnitSet OpenBabel::FindStereogenicUnits | ( | OBMol * | mol, |
| const std::vector< unsigned int > & | symClasses | ||
| ) |
Find the stereogenic units in a molecule using a set of rules.1
The potential stereocenters are identified first. A potential tetrahedral stereogenic atom is any atom meeting the following criteria:
Nitrogen is treated as a special case since the barrier of inversion is low in many cases making the atom non-stereogenic. Only bridge-head nitrogen atoms (i.e. nitrogen has 3 neighbors in rings) will be considered stereogenic.
Potential stereogenic double bonds are identified using another set of simple criteria:
True stereocenters (i.e. stereocenters with topologically different ligands) are identified first. For tetrahedral stereocenters, true stereocenters will have 4 different neighbor atom symmetry classes and this can be expressed using T1234 to classify these stereocenters. For stereogenic bonds, a similar classification C12 can be used but both begin and end atom have their own classification and the bond is only a true stereocenter if both atoms are C12.
Para stereocenters are all stereocenters where there are at least two equivalent neighbor atom symmetry classes. These are T1123, T1112, T1111 and T1122 for tetrahedral stereocenters and C11 for double bonds. To determine which of the remaining potential stereocenters really are stereocenters, a set of rules is used.1
Rule 1 is applied recusively:
All rings are merged "mergedRings". A merged ring is simply a fragment consisting of all atoms of a ring system (bridged, spiro, adjacent, ...). If two rings in the SSSR set share an atom, they are merged.
Each merged must at least have two para-stereocenters (or 1 true + 1 para) in order for the para-stereocenter to be valid. This is repeated until no new stereocenters are identified.
rule 1a for double bonds:
rule 1b for tetracoord atoms:
Rules 2 and 3 are applied sequential (i.e. only once).
Rule 2a for tetracoordinate carbon:
Rule 2b for tetracoordinate carbon:
Rule 3 for double bonds:
Reference:
[1] M. Razinger, K. Balasubramanian, M. Perdih, M. E. Munk, Stereoisomer
Generation in Computer-Enhanced Structure Elucidation, J. Chem. Inf.
Comput. Sci. 1993, 33, 812-825
Referenced by OpenBabel::CanonicalLabels().
| OBStereoUnitSet OpenBabel::FindStereogenicUnits | ( | OBMol * | mol, |
| const std::vector< unsigned int > & | symClasses, | ||
| const Automorphisms & | automorphisms | ||
| ) |
Find the stereogenic units in a molecule making use of the automorphisms.
The potential stereocenters are identified first. A potential tetrahedral stereogenic atom is any atom meeting the following criteria:
Nitrogen is treated as a special case since the barrier of inversion is low in many cases making the atom non-stereogenic. Only bridge-head nitrogen atoms (i.e. nitrogen has 3 neighbors in rings) will be considered stereogenic.
Potential stereogenic double bonds are identified using another set of simple criteria:
Once the potential stereocenters are found, the automorphisms are the key to identifying real stereogenic units. Automorphisms can be seen as permutations that permutate a graph back to the same graph. Such a permutation can only exchange atoms with the same symmetry class and it follows that the use of automorphisms takes symmetry into account. The definitions below use a concept where the automorphisms cause inversions of configuration to potential stereocenters. Such an inversion occurs whenever an automorphism exchanges two equivalent (i.e. with the same symmetry class) neighbor atoms attached to the potential stereogenic unit.
The actual number of configurations needed for the ligands depends on the classification (i.e. T1234, T1123, ...) of the stereo center. These classes reflect the symmetry classes of the neighbor atoms of the center.
For stereogenic bonds, there is only one case but both begin and end atom have to be checked.
These criteria are analogous to the rules from the Razinger paper on stereoisomer generation. Since the existance of stereocenters can depend on the existance of other stereocenters (in the ligands), the stereocenters are found by iterating until no new stereocenters are found.
Reference:
[1] M. Perdih, M. Razinger, Stereochemistry and Sequence Rules:
A Proposal for Modification of Cahn-Ingold-Prelog System,
Tetrahedron: Asymmetry, 1994, Vol. 5, No. 5, 835-861
| ostream& std::operator<< | ( | ostream & | out, |
| const OpenBabel::OBTetrahedralStereo & | ts | ||
| ) |
OBTetrahedralStereo::Config cfg; cfg.center = 0; cfg.towards = 4; cfg.refs = OBStereo::MakeRefs(1, 2, 3); cfg.winding = OBStereo::AntiClockwise; cfg.view = OBStereo::ViewTowards; OBTetrahedralStereo ts(mol); ts.SetConfig(cfg) cout << "ts = " << ts << endl; // output OBTetrahedralStereo(center = 0, viewTowards = 4, refs = 1 2 3, anti-clockwise)
| ostream& std::operator<< | ( | ostream & | out, |
| const OpenBabel::OBTetrahedralStereo::Config & | cfg | ||
| ) |
OBTetrahedralStereo::Config cfg; cfg.center = 0; cfg.towards = 4; cfg.refs = OBStereo::MakeRefs(1, 2, 3); cfg.winding = OBStereo::AntiClockwise; cfg.view = OBStereo::ViewTowards; cout << "cfg = " << cfg << endl; // output OBTetrahedralStereo::Config(center = 0, viewTowards = 4, refs = 1 2 3, anti-clockwise)